Overview of predicted results
Overview of the results
| Contig_ID | Contig_def | CRISPR array number | Signature genes | Novel effector protein number |
|---|---|---|---|---|
| QEUN01000024 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_6810, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000010 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_173, whole genome shotgun sequence | 2 CRISPRs | 0 | |
| QEUN01000028 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_11528, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000014 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_278, whole genome shotgun sequence | 1 CRISPR | 1 | |
| QEUN01000006 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_110, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000003 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_40, whole genome shotgun sequence | 3 CRISPRs | 0 | |
| QEUN01000018 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_482, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000022 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_1545, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000007 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_115, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000027 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_11379, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000029 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_57135, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000015 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_365, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000002 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_35, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000012 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_204, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000025 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_7526, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000005 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_82, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000020 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_870, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000013 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_237, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000001 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_15, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000023 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_3419, whole genome shotgun sequence | 1 CRISPR | 0 | |
| QEUN01000019 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_655, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000008 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_138, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000011 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_186, whole genome shotgun sequence | 1 CRISPR | 0 | |
| QEUN01000026 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_9253, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000016 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_370, whole genome shotgun sequence | 1 CRISPR | 0 | |
| QEUN01000021 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_1357, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000009 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_143, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000017 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_372, whole genome shotgun sequence | 0 CRISPR | 0 | |
| QEUN01000004 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_49, whole genome shotgun sequence | 0 CRISPR | 0 |
1. QEUN01000014
| Group ID | Candidate ID | Protein size | Protein distance | Strain info | CRISPR_ID | Homolog(s) |
|---|---|---|---|---|---|---|
| 1 | QEUN01000014.1|RIJ98969.1|51295_53869_+|hypothetical-protein | 857 aa | 3 | Armatimonadetes bacterium isolate ATM2 J3_scaffold_278 | QEUN01000014_1 | 1 |
Click the colored protein region to show detailed information
| Group ID | Candidate ID | Protein size | Protein distance | Strain info | CRISPR_ID | Homolog(s) |
|---|---|---|---|---|---|---|
| 1 | RHKV01000006.1|KAA0225664.1|[187352:189956](+)|hypothetical_protein | 867 aa | 3 | Armatimonadetes bacterium isolate ARM2 N2_1.scaffold_15, whole genome shotgun sequence | RHKV01000006_1 | Homolog protein of candidate |
Click the colored protein region to show detailed information
Novel class2 CRISPR loci
| CRISPR_ID | CRISPR_location | CRISPR_type | Repeat_type | Spacer_info | Cas protein info |
|---|---|---|---|---|---|
| QEUN01000014_1 | 56374-57741 | Unclear |
NA
|
18 spacers
|
cas1,cas2,cas4 |
| RHKV01000006_1 | 192391-195093 | Unclear |
NA
|
36 spacers
|
cas1,cas2,cas4 |
You can click texts colored in the table to view more detailed information
Novel effector protein sequence
>QEUN01000014.1|RIJ98969.1|51295_53869_+|hypothetical-protein MSPLERSLRKVGENRLERLRVREEKIRKHIEQHPRGKNDHQALHFLLHQIEVERNDLYRNLKDPEYVPKPAKQRRERRQINVAKPPTRPKKEKGPQPESTKYVIRPPVPGKNLPAFASKYEARDTRDDSYQDGRSWTSAPYVEVELPILGADKVIQKLMKFVQKDERSIVRDWATKTYSSIEAAREALLVGAQVSEDVSVWRGLLAETKNAQNFAALSDDQIEAAMSKEAKGADLRPRRAALLVAQRHWVDQTVKAIKESAPSGVDKDTLDRRLRAGLRGFHTAANSGKHTNPQFPYLTAEKPVVPMESVVQSVLAFLDDPDDQRYTKDKEDDKKRHRVTVLQKELGKARPRKRLELQTPKWAGRPTVKGTISKRRDAALVWDTSKEANGLCLALPIGGMPKIDVEQFIYQDGTSLLSDCQIASKTTKKGAACAVLPLKPKHDFLRWFTKHVENHNPDAPLERRCLHNTTQFVIVDPEGPRPRLFVRPVFKFYDPGKTVPNTHETWKKPDCRYLVGIDRGINYVLRAVVVDTEEKKVIADIGLPGRKHEWRMIRDEIAYHQQMRDLARNTGKHASVVAKHVRALALARKKDRALGKFATVEAVAELVKKCEQDYGSGNYCFVLEDLDMGAMNLKRNNRVKHMAVMEEALVNQMRKQGYAYDGRRGRVDGVRHEGAWYTSQVSPFGWWAKRDEVEEAWKRDKTRPIGRKVGNWYEMPEPGQDGDRPDTYRKGYWSKPKNAEGKPYGRNRFSVEPGDEKPDAERRFCWGSELFWDPNVKSFKGKEFPEGVVLDADFVGALNIALRPLVNDGQGKGFKAEDMAREHTILNPQFKIACQIPVYEFVEEDGDKWAALRRIML >|RHKV01000006.1|KAA0225664.1|[187352:189956](+)|hypothetical_protein|149|867|GCA_008363305.1_ASM836330v1 MAKATKEVKSKRVEALRQVAYQRLERLERKAQKIGAHLRKPGKAADLQSLHYLLHKVEVEYHDIARNLEKDPTWTPKPKMRREKRAIVPESGPAAPLPTTAKGEPGRPANRHIPPPVPLDSARIPEDQQSMGQGSGGRSWCSAPFVEVKLPPTQWSNVREKLLKFRIEDDADIVRRWAEAKFGSIETARDGLRASAEIGTSPDVWRSFISRAISNGKKDFEPLLSLDDDELTADATAERVVRRWHQIDWVGRMLDSILETVPSGVSKDTFRSRVESRLKTFHSSVNSFELKKRKDGTVERKRKHTNPQFPYLSPSAVSIDPDVVTMEAVELLQMQPEERFAKDPNDANGRMRLRVLQAELGKARREALGRRGEKAPPWSGRKVFRGTTTRKREACLVWDKEAQADGLYFALVMSGGPKIDDKRFVYMDGQPLQSDWQLHNGVAGKAKSCRAMPLILKHDFLRWYHRHIKNHDVNAPLEKRCVHTTTQFVFVEPDEKKGLQPRLFIRPVFKFYDPVYEVPDSHSIDKKPDCRYLIGIDRGVNYPYRAAVYDCETNSIIADKFVDGRKADWERIRNELAYHQRRRDLLRNSRASSAAIQREIRAIARIRKRERGLNKVETVESIARLVDWAEENLGKCNYCFVLEDLSSNLNLGRNNRVKHIAAIKEALINQMRKRGYRFKKSGKVDGVREESAWYTSAVAPSGWWAKKEEVDGAWKADKTRPLARKIGSYYCCEEIDGLHLRGVLKGLGRAKRLVLQSDDPSAPTRRRGFGSELFWDPYCTELCGHAFPQGVVLDADFIGAFNIALRPLVREELGKKAKAVDLADRHQTLNPTVALRCGVTAYEFVEVGGDPRGGLRKILLNPAEAVI
Repeat information
| CRISPR_ID | CRISPR_type | repeat_type | Consensus_repeat | Method |
| QEUN01000014_1 | Unclear | NA | GTCGCAGGGGATCAAGAACGCTCTTAGGGAATGAAA | PILER-CR |
| QEUN01000014_1 | Unclear | NA | GTCGCAGGGGATCAAGAACGCTCTTAGGGAATGAAAG | CRISPRCasFinder |
| QEUN01000014_1 | Unclear | NA | GTCGCAGGGGATCAAGAACGCTCTTAGGGAATGAAAG | CRT |
| RHKV01000006_1 | Unclear | NA | GGCGCAGGACAAAATGCACACTCTAAAGGAATGAAAG | PILER-CR |
| RHKV01000006_1 | Unclear | NA | GGCGCAGGACAAAATGCACACTCTAAAGGAATGAAAG | CRISPRCasFinder |
| RHKV01000006_1 | Unclear | NA | GGCGCAGGACAAAATGCACACTCTAAAGGAATGAAAG | CRT |
Spacer information
>1.1|56410|41|QEUN01000014|PILER-CR GACGGGCAGTATCGGAGGGCGCGGGAGAGGTGCCAGCCTTC >1.2|56487|38|QEUN01000014|PILER-CR GCGTTGCAGGCAATGATCTCTGCAACTGGCCGTCGAAT >1.3|56561|36|QEUN01000014|PILER-CR GGGAGTGGGTGCGACTTAAAGCGTTTGACGGTGGCT >1.4|56633|37|QEUN01000014|PILER-CR GAAACAAAGACGAATTGCCTGGACGCAGGCAAAATAC >1.5|56706|39|QEUN01000014|PILER-CR GATTGCGACAGCCTCGCCTCATTCCAGGCCGACAGGCTG >1.6|56781|38|QEUN01000014|PILER-CR GATCGTTCCTGCACGAGAGCGGAGTGAGTCGCCGGGGG >1.7|56855|38|QEUN01000014|PILER-CR GGATTACCTAACCACCCCCAGCAGGGCTTCCGGCCCTG >1.8|56929|40|QEUN01000014|PILER-CR GGGAGAGACGGGCAGGGAGAATGAACAATGAAAGCGATCC >1.9|57005|34|QEUN01000014|PILER-CR GCTCGCCCAACCCTCGGACAGGTTGGGTGGCGGG >1.10|57075|36|QEUN01000014|PILER-CR GTTGAACCCGCCAAAGCAGTTCAGCCACGCCGCCAG >1.11|57147|38|QEUN01000014|PILER-CR GTATTCCACGCAGCACATCGGAGAACTTGCGTTTCGGT >1.12|57221|38|QEUN01000014|PILER-CR GTGCGTAGTGCGAGCGGATCTATCCGAGGTTGACCGGA >1.13|57295|39|QEUN01000014|PILER-CR GGTATGCTCCAAGACCTTCGTCGTCGGGTTCCAGGCCAT >1.14|57370|37|QEUN01000014|PILER-CR GTGTAATTCTGATACAGGCGCGACCTCACGAGCACCC >1.15|57443|37|QEUN01000014|PILER-CR GCATTCTGCGCGGCCTATACATCGAATACCTGCGAAC >1.16|57516|40|QEUN01000014|PILER-CR GATTGCCTTCAGCCGCTTAGCCATTCGCTCGGCATAGCAA >1.17|57592|39|QEUN01000014|PILER-CR GTTTGCCGGCCGAGACGTGCCACCGCCTAGTCAGGCGAT >1.18|57667|38|QEUN01000014|PILER-CR GAGGTTTCGGGCCTGGAATACCCGGCACCCGGTCGGAT >1.19|56411|40|QEUN01000014|CRISPRCasFinder,CRT ACGGGCAGTATCGGAGGGCGCGGGAGAGGTGCCAGCCTTC >1.20|56488|37|QEUN01000014|CRISPRCasFinder,CRT CGTTGCAGGCAATGATCTCTGCAACTGGCCGTCGAAT >1.21|56562|35|QEUN01000014|CRISPRCasFinder,CRT GGAGTGGGTGCGACTTAAAGCGTTTGACGGTGGCT >1.22|56634|36|QEUN01000014|CRISPRCasFinder,CRT AAACAAAGACGAATTGCCTGGACGCAGGCAAAATAC >1.23|56707|38|QEUN01000014|CRISPRCasFinder,CRT ATTGCGACAGCCTCGCCTCATTCCAGGCCGACAGGCTG >1.24|56782|37|QEUN01000014|CRISPRCasFinder,CRT ATCGTTCCTGCACGAGAGCGGAGTGAGTCGCCGGGGG >1.25|56856|37|QEUN01000014|CRISPRCasFinder,CRT GATTACCTAACCACCCCCAGCAGGGCTTCCGGCCCTG >1.26|56930|39|QEUN01000014|CRISPRCasFinder,CRT GGAGAGACGGGCAGGGAGAATGAACAATGAAAGCGATCC >1.27|57006|33|QEUN01000014|CRISPRCasFinder,CRT CTCGCCCAACCCTCGGACAGGTTGGGTGGCGGG >1.28|57076|35|QEUN01000014|CRISPRCasFinder,CRT TTGAACCCGCCAAAGCAGTTCAGCCACGCCGCCAG >1.29|57148|37|QEUN01000014|CRISPRCasFinder,CRT TATTCCACGCAGCACATCGGAGAACTTGCGTTTCGGT >1.30|57222|37|QEUN01000014|CRISPRCasFinder,CRT TGCGTAGTGCGAGCGGATCTATCCGAGGTTGACCGGA >1.31|57296|38|QEUN01000014|CRISPRCasFinder,CRT GTATGCTCCAAGACCTTCGTCGTCGGGTTCCAGGCCAT >1.32|57371|36|QEUN01000014|CRISPRCasFinder,CRT TGTAATTCTGATACAGGCGCGACCTCACGAGCACCC >1.33|57444|36|QEUN01000014|CRISPRCasFinder,CRT CATTCTGCGCGGCCTATACATCGAATACCTGCGAAC >1.34|57517|39|QEUN01000014|CRISPRCasFinder,CRT ATTGCCTTCAGCCGCTTAGCCATTCGCTCGGCATAGCAA >1.35|57593|38|QEUN01000014|CRISPRCasFinder,CRT TTTGCCGGCCGAGACGTGCCACCGCCTAGTCAGGCGAT >1.36|57668|37|QEUN01000014|CRISPRCasFinder,CRT AGGTTTCGGGCCTGGAATACCCGGCACCCGGTCGGAT >GCA_008363305.1_ASM836330v1|1.1|192428|36|RHKV01000006 GGTCACGGCCACGGTGAGCGCGACGGACGTGAGCTC >GCA_008363305.1_ASM836330v1|1.2|192501|34|RHKV01000006 GAGGGCTGGAAACTCGAAATCCTCGACACGGGCG >GCA_008363305.1_ASM836330v1|1.3|192572|37|RHKV01000006 GAAAAGAATCGAGACGGCGCAAAGCCGAAAGAATACT >GCA_008363305.1_ASM836330v1|1.4|192646|37|RHKV01000006 GGTTCTGATAGCCAAGTAGTGCCTCGATTCCCACCCT >GCA_008363305.1_ASM836330v1|1.5|192720|38|RHKV01000006 ATCAGCTCGAGGGCGTTCGCAAGCGCGACCGCTTCCTC >GCA_008363305.1_ASM836330v1|1.6|192795|37|RHKV01000006 ACGGAGAGTGGATGCTCTTGGCACGAGAGCGCTGGCA >GCA_008363305.1_ASM836330v1|1.7|192869|38|RHKV01000006 ACACGACGCGCGGGTGAGCCGCGAGGAGTTCCGGCTGC >GCA_008363305.1_ASM836330v1|1.8|192944|37|RHKV01000006 AGGCAGATGCGCGGCGGACCATGATCGCGACACTCCT >GCA_008363305.1_ASM836330v1|1.9|193018|37|RHKV01000006 CTCGTGGAAGTTCCGGAAGCGCTCTTCAGTAGAGAGC >GCA_008363305.1_ASM836330v1|1.10|193092|37|RHKV01000006 ACAACGCAGCGTCTCGCAACAGGCCGTATCTCGCAGT >GCA_008363305.1_ASM836330v1|1.11|193166|35|RHKV01000006 GAGCAGAACGCGGCGCAATACGGGAGGGGCGGACC >GCA_008363305.1_ASM836330v1|1.12|193238|37|RHKV01000006 GGATATGTTTCCGTGAACGCCGGTCGCTCGGACGCCG >GCA_008363305.1_ASM836330v1|1.13|193312|41|RHKV01000006 TCTTTTAGCGTCGAAAAACCCCGACATTTTTCGCCAAAACC >GCA_008363305.1_ASM836330v1|1.14|193390|38|RHKV01000006 TGCATAACTCTACCTCGCTAATAGCGCGTGAACGGAAA >GCA_008363305.1_ASM836330v1|1.15|193465|38|RHKV01000006 GAAGTACAGGGCGTCATTCGTGTCGAACGGATCCGGCA >GCA_008363305.1_ASM836330v1|1.16|193540|37|RHKV01000006 ACACTCGGCGCGCACTTGCAAGATGAAAGAGCCCCCA >GCA_008363305.1_ASM836330v1|1.17|193614|37|RHKV01000006 CGAATCGGCCACGTCCACGGCCCCTCAGACGGGTCGA >GCA_008363305.1_ASM836330v1|1.18|193688|39|RHKV01000006 AGCCGAGCGTCAGGGGTTTCATTCCGTCTCTCCTTTCGC >GCA_008363305.1_ASM836330v1|1.19|193764|37|RHKV01000006 TTGCAAGTCTCATTCGATTGCCTCCATGATTGCTTGA >GCA_008363305.1_ASM836330v1|1.20|193838|37|RHKV01000006 CGGCTACGCACGGATGGACTCCGCACGCGTAATCTCT >GCA_008363305.1_ASM836330v1|1.21|193912|36|RHKV01000006 TAGTCGAATGCGTTTTGATCGCGAATGCGCGTAACG >GCA_008363305.1_ASM836330v1|1.22|193985|38|RHKV01000006 CCAGCCACTTCCATATCCGTCGATCGCGCGCGAGCAGT >GCA_008363305.1_ASM836330v1|1.23|194060|36|RHKV01000006 GTAAAGGAGATCGTGGCAGCCGCGACCGCGCTCTTG >GCA_008363305.1_ASM836330v1|1.24|194133|37|RHKV01000006 TGTGATTGCAGGTCCGAGTGCATCGCTCGCGCATCCT >GCA_008363305.1_ASM836330v1|1.25|194207|36|RHKV01000006 CGCGTCTCTTATAGGGATCAGTCGGAATCCTCATTA >GCA_008363305.1_ASM836330v1|1.26|194280|36|RHKV01000006 CAACTGGATGGTCAGAATGCGGTCGTACTCTGTGGG >GCA_008363305.1_ASM836330v1|1.27|194353|35|RHKV01000006 TGACGACGACGTTCGGCTCGCGATTCTGAATCGCG >GCA_008363305.1_ASM836330v1|1.28|194425|38|RHKV01000006 TTACCAAAACCGAAACCATTATTGGGAATGGGGAAAAC >GCA_008363305.1_ASM836330v1|1.29|194500|37|RHKV01000006 ACCCGTCGGTGAACTCGACGAACAAGTCCTCGATGTC >GCA_008363305.1_ASM836330v1|1.30|194574|38|RHKV01000006 GAGAAGACCCCCGCGAGGCGAAGAACCTGGGCCATCTG >GCA_008363305.1_ASM836330v1|1.31|194649|38|RHKV01000006 GGATCCTCGAGCCTGAGCAGTAATGGACTGGTCGGAGG >GCA_008363305.1_ASM836330v1|1.32|194724|38|RHKV01000006 GGTCCGCTGGGCGCTGTCGCACTACGGTCGCTGCCGGC >GCA_008363305.1_ASM836330v1|1.33|194799|35|RHKV01000006 ACTCCAGGGCCTCCCTGTTGGTGAAGAACAGATAG >GCA_008363305.1_ASM836330v1|1.34|194871|37|RHKV01000006 TCTTATCTGCCTCCGGCATTTATTTCTTCTTGGGGCG >GCA_008363305.1_ASM836330v1|1.35|194945|37|RHKV01000006 TCCCTGAGCCCTTGCAAGTCCAGTGAGACGTTGTCGA >GCA_008363305.1_ASM836330v1|1.36|195019|37|RHKV01000006 TTGTAGACGACGTTCGAGTAGACGCGCGAGCCCGGCA
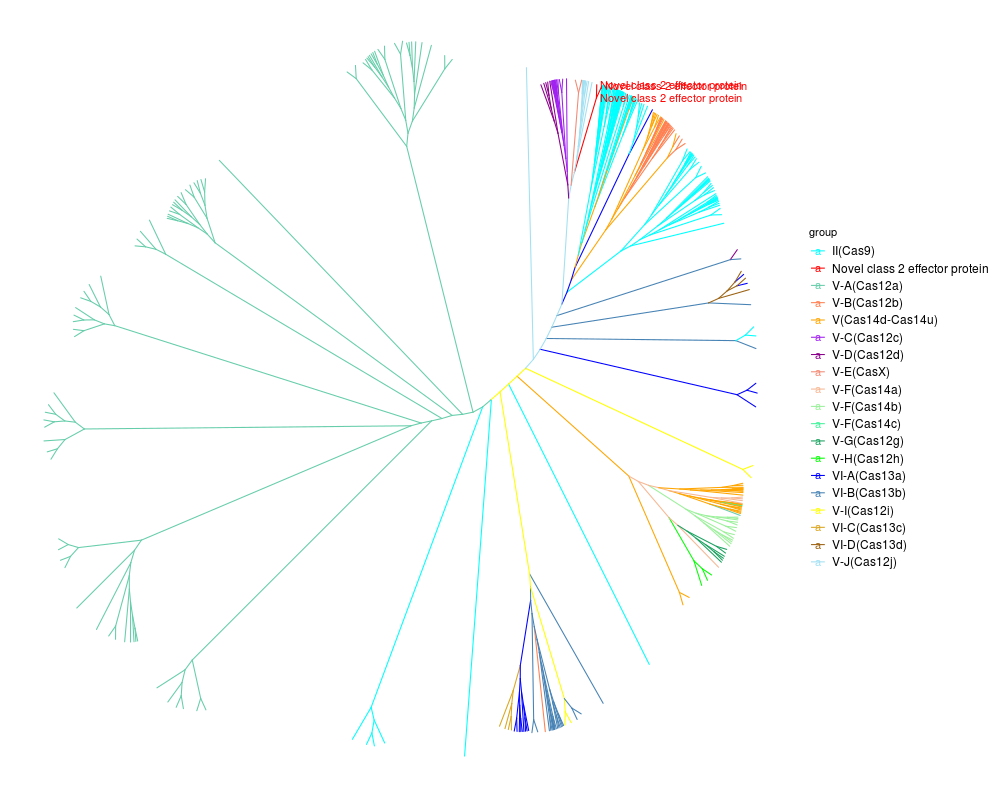
Maximum likelihood phylogenetic tree of class 2 effector proteins